Possible bug in bayesian paired wilcoxon?
Hi there,
I have been using the bayesian version of the paired wilcoxon test to analyze some of my data and have been wondering if there might be a bug based on the output I got. More specifically, I simulated two groups in R with rnorm: s_test (n=100, m=20, sd=5), c_test (n=100, m=25, sd=5) and analyzed them in JASP. I used one-sided tests (Measure 1 < Measure 2) and conducted a t-test and a Wilcoxon.
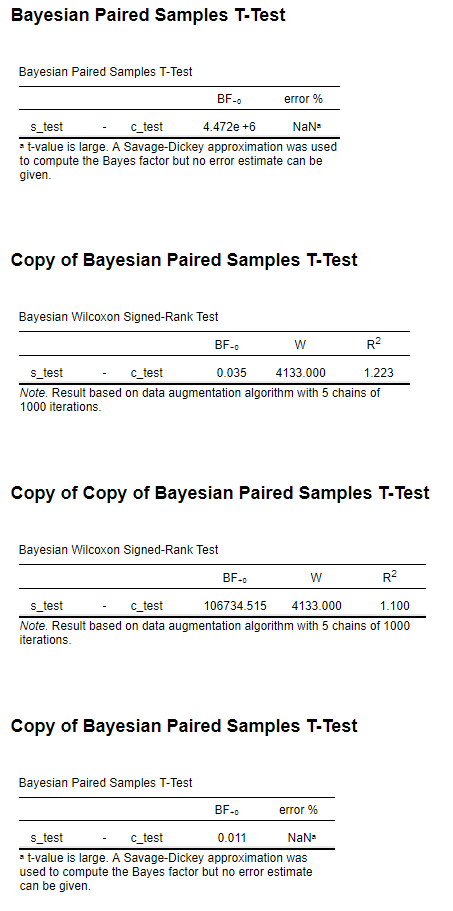
The t-test gives me a BF-0 4.472e+6 and the Wilcoxon a BF-0 0.035 (see image). I have a feeling that the alternative hypothesis for the Wilcoxon is coded inversely (as Measure 1 > Measure 2) as I get a more realistic result for the Wilcoxon when I switch the alternative hypothesis (BF+0 106734).
So here the question: Is it possible that there is a bug, or am I missing something important?
I have also included my simulated data if anyone wants to replicate the analysis.
Best,
yvonne


Comments
Hi Yvonne,
Thanks for reporting this! We are just about to do a new version, so this is perfect timing. I'll forward this to Johnny. For future occasions, it would be great if you could use our GitHub page (for details see https://jasp-stats.org/2018/03/29/request-feature-report-bug-jasp/).
Cheers,
E.J.
Dear E.J.,
thank you! I will use GitHub in the future.
Best,
yvonne