JASP Results not comparable to MATLAB or R
Hi there,
so I am running the same Data through a GLMM in JASP, MATLAB & R to get an overview/confidence in my results. Its a simple Generalized Linear Mixed effect model on reaction times being influenced by the stimulus type and the movement type that was to be performed.
While MATLAB and R have congruent results JASP shows some oddities. As to be seen in the attached images. Especially odd is the difference in DF, where MATLAB consistently choose the number of trials (7063) and JASP chooses the Number of participants (~23) for the intercept. This is very odd and does not react to any changes in the model.
If you have any idea why this is the case, please let me know!


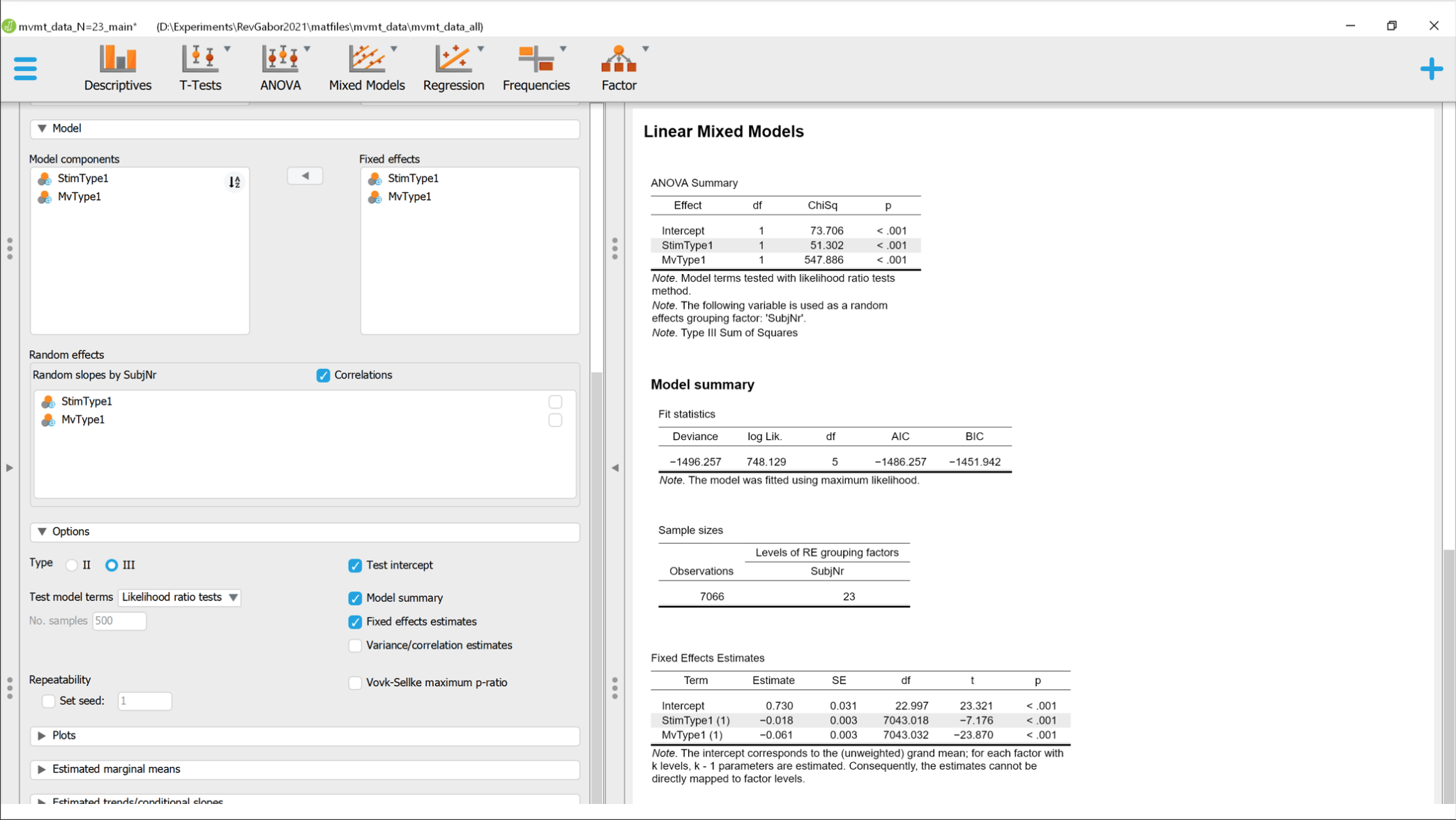

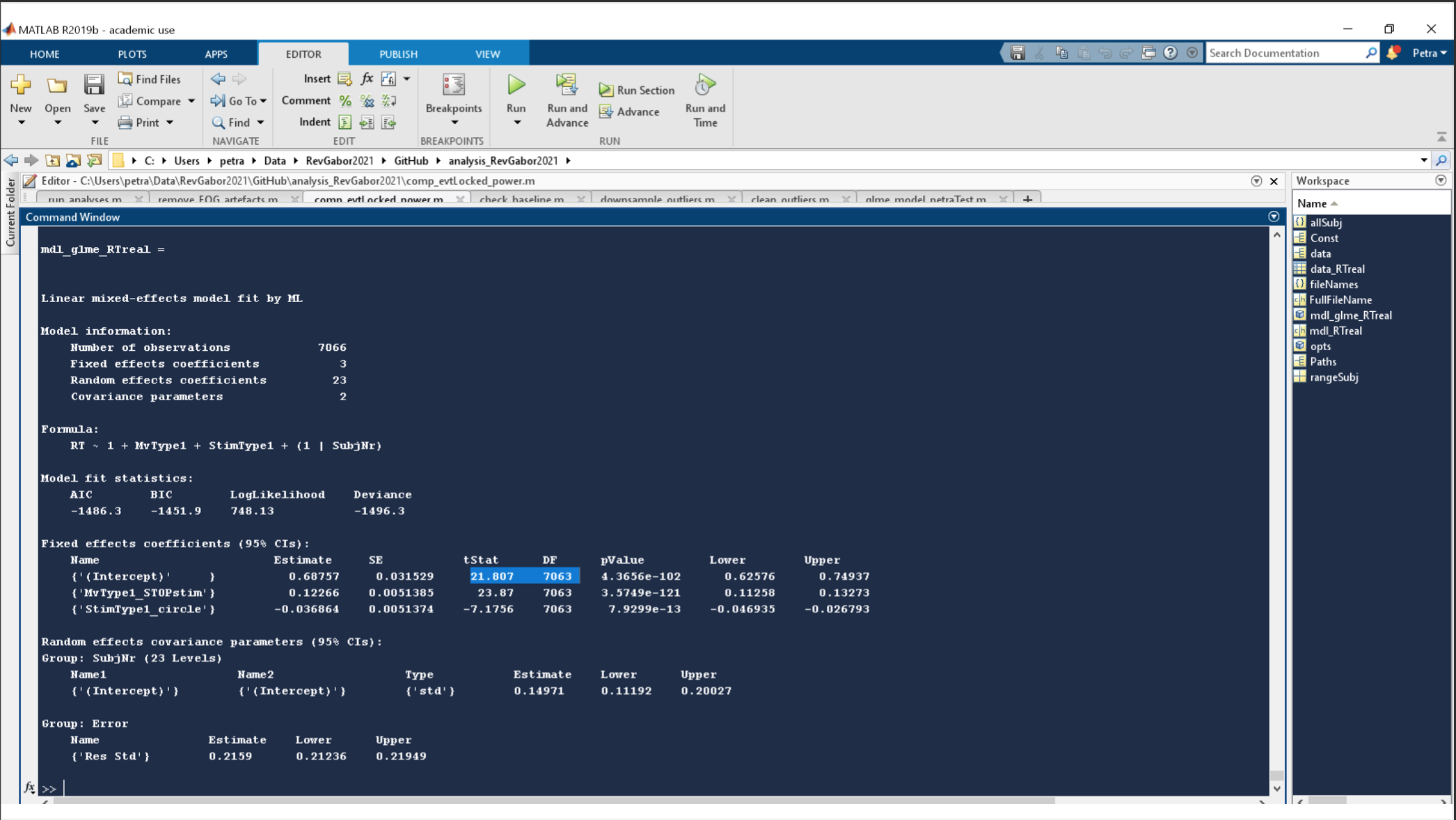
Comments
Two things are going on: