Issues with Generalized Mixed Models procedure in JASP
Hi everyone,
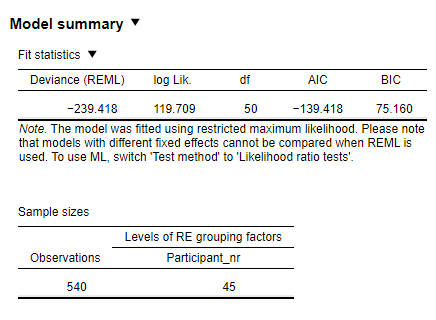
I am a neuroscientist trying to run mixed models in JASP on psychophysics data. I am using a Gaussian distribution and Identity link function to model the data and would like to compare models that have different fixed effects using ML. To achieve this, I used the JASP frequentist Generalized linear mixed models procedure. However, I am unsure which method (ML or REML) JASP uses for the Generalized Mixed Models function by default. Namely, when I run my analyses, the "Test method = Likelihood ratio test" option is ticked. Nonetheless, despite that, the output still says that the models are estimated with REML and I am once again directed to choose the "Test method = Likelihood ratio test" option, although it is already chosen (see an example below):
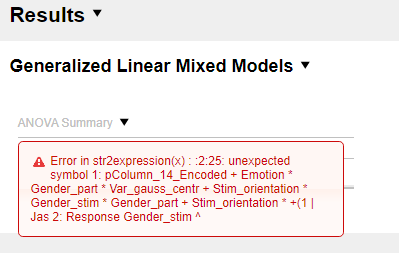
Therefore, could you please clarify whether ML or REML is used by default for the Generalized Linear Mixed Models procedure and how one can switch between ML and REML testing in JASP? Additionally, assuming that ML is in fact used for estimating the models, I performed some model reductions for the fixed effects. However, I receive this error message after removing a few non-significant interactions:
It is unclear to me what the issue is. Do you perhaps know what goes wrong? Finally, I would like to know which covariance structure JASP uses for the Generalized Linear Mixed Models procedure and where one can change the structure? Thank you for your assistance in advance!


Comments
Seeing as you're using Gaussian with identity link, have you tried using lineard mixed instead of generalized linear mixed? (Linear mixed is Gaussian with identity link)
Hi patc3, Thank you for your reply. I gave it another try with Linear mixed models as you suggested, however without success. Once I remove the last 4-way interaction, I again receive this error message:
I do have a covariate in the model, in case that is relevant information. These are my variables:
• DV – Response (0- unseen, 1 – seen) – data has been averaged across 2 runs and blocks, so the values are in decimals now
• IV1 – Emotion (threat, fear, sadness)
• IV2 – Stimulus orientation (upright/inverted)
• IV3 – Stimulus gender (male, female)
• IV4 – Participant gender (male, female)
• Covariate – White noise centered (the mean was removed)
DV = dependent variable, IV = independent variable
I've forwarded this to our expert
E.J.
This seems to be an issue with the maximum length of R formula -- it cannot be longer than 500 characters. In such a case, I would recommend shortening variable names (e.g., Emotion -> Em, Stimulus orientation -> StOr, etc.). In case that does not solve the issue, let us know, please.
(This suggestion will now appear instead of the error message)
Cheers,
Frantisek