JASP Omega different from RStudio
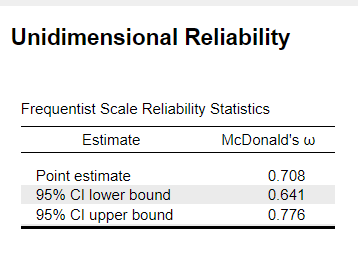
I get an Omega value of 0.708 in JASP with the function Unidimensionality Reliability, but a value of 0.74 in RStudio.
I can't see the R code for this function in JASP, but the R Code I used in omega(df.items, nfactors=1). I attached the 4-item dataset.



Comments
Hi,
the difference stems from the psych package in R using the data correlation matrix to find the factor loadings to compute omega, and in JASP we use the covariance matrix, because we deem it more informative.
Julius
That makes sense. Thank you!
So basically one is standardized and the other isn't?
Yes, that's how I understand it.
One can add the following R code to give the unstandardized (from covariance matrix): covar = TRUE.
To be exact, to get the same value as JASP: omega(Dataset, nfactors = 1, fm = "ml", covar = TRUE).
Exactly. We have had a discussion with Klaas Sijtsma about this, and we came to the conclusion that through the standardisation differences in the variances are lost. It is true however, that we offer standardised alpha - because it is somewhat of a convention - and in the Bayesian version we offer all coefficients standardized. So eventually, we should probably also offer all coefficients standardized for the frequentist reliability as well.