Parallel analysis for EFA in JASP
Hi,
I wanted to ask which underlying R function JASP uses to run the parallel analysis in order to quantify the number of factors for an exploratory factor analysis?
I ran the same EFA in JASP and R using the fa.parallel function (psych package). With the fa.parallel function, I get 6 factors no matter which factor method I use (minres, ml, wls, gls, pa). However, JASP outputs 10 factors when I use the parallel analysis option to extract the number of factors.
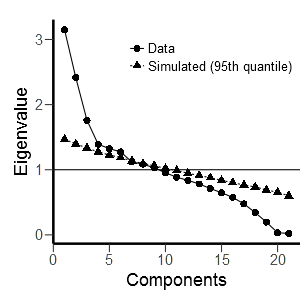
Looking at the scree plot, I can see that one could argue for two different points of inflexion, yielding either 6 or 10 factors. However, based on the scree plot, I would think that 6 factors are the more straightforward solution and I am puzzled that the parallel analysis in JASP yields a different result. Therefore, I would really appreciate your help!


Comments
I'll ask our EFA expert. Sorry for the tardy response, this slipped through the cracks...more to follow
Cheers,
E.J.
Probably too late but in case other people run into this issue too: Note that the fa.parallel function will by default run an EFA to extract a one factor solution to obtain the communality estimates and compute the eigenvalues from the correlation matrix with the communality estimates as diagonal. This is done both for the real correlation matrix as well as for the simulated data. This is not the usual implementation of parallel analysis, where eigenvalues are found from the correlation matrix (with the diagonal replaced by the squared multiple correlations in the case of PA-PAF), and can lead to different results. You can also run the usual parallel analysis procedure in fa.parallel by setting SMC = TRUE. Probably fa.parallel will then also yield 10 factors.
Hmmm I see our expert has not responded yet...I'll prompt him :-)
Sorry for the late response!
What you find is strange, because in JASP the EFA parallel analysis is just a vanilla implementation of
psych::fa.parallel(dataset)Looking at your plot, it could be that this difference is due to chance anyway (because the simulated data can be a little different every time), even though you tried multiple times. The simulated-observed lines almost overlap at 6 factors.
In any case, we have to remind ourselves that this is just a tool for determining the number of factors. If you look at a plot like this where the differences are minimal and you think 6 factors is better (also taking into account, for example, subject knowledge), then I suggest using 6 factors.
Erik-Jan
Thank you so much for the explanation and help!
To follow up on this older thread:
I have the same issue in that JASP always suggests a different number of factors compared to R in parallel analysis, irrespective of which fa.parallel() settings I use, i.e., SMC = TRUE does not make a difference. I have tried this for 3 different combinations of questionnaires (different combinations of items from the same data set in this case), and I have repeated the parallel analysis several times.
Could there maybe be a different explanation for why this happens? The factor loadings are identical as expected when I use "minres" for both.
As an example, R suggests 3 whereas JASP suggests 1 factor here. I can see how both make sense, but this happens for any combination of items.
Any help is appreciated!
Does anyone know whether the question above can really just be due to differences in simulated data? I think the question might have slipped through the cracks.
As it is a consistent difference between R and JASP for all my data sub-sets and has happened before to other users I find that hard to believe. For the EFA that I will run next week, I just would like to make sure I can trust either R or JASP.
Thank you in advance!
(Adding info here for readers) The source of the discrepancy has been found and fixed: the number of principal components rather than the number of factors was being retrieved by JASP from the parallel analysis. See the corresponding pull request on GitHub