Implausible trajectories from Qualtrics
Hello,
Thanks in advance for any advice. I've recently collected some Mousetracking data using qualtrics (following this code https://osf.io/st2ef/). I've adapted their dataprep file in order to be able to import it as a mousetrap object
When I try and align (mt_data <- mt_align_start(mt_data)) my data it seems to put it upside down and I get this warning
Warning message:
In mt_reshape(data = data, use = use, use2 = use2, use2_variables = use2_variables, :
For some trials in data[[use2]], no corresponding trials in data[[use]] were found.
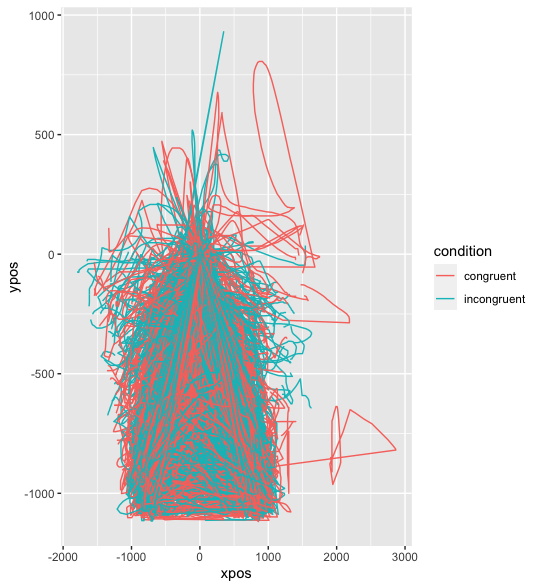
The thing is is that there are these really odd trajectories that are outside of the window (i.e. impossible to be real). This is not fixed by only keeping items with 2+ observations (> mt_example <- mt_subset(mt_example, nobs>2)) I am able to subset the data depending on xpos_max (mt_example <- mt_subset(mt_example, xpos_max<=1000, check="measures") and then align the data and this makes more sense
However there are still some implausible trajectories (i.e. >+1000/ <-1000 xpos) and mt_example <- mt_subset(mt_example, xpos_min>=0, check="measures") doesn't remove y trajectories with less than 0 and they don't all end where the left/right buttons were
When I try and remap my data symmetrically it throws this error:
mt_data <- mt_remap_symmetric(mt_data)
Error in if ((remap_xpos == "left" & trajectories[i, nlogs, xpos] > 0) | : argument is of length zero
When I subset my browser version or into individual participants sometimes I can remap symmetrically and some participants have ideal trajectories (i.e. all plausible; start and finish in same location and don't go outside possible window), some have the odd implausible trajectory and some are completely implausible (upside down and ending where ever on the screen not the designated end button).
I'm wondering if it is converting the X and Y co-ordinates from relative timestep increments to position on screen? Any advice would be greatly appreciated


Comments
Hi,
I think that there are several questions and issues to discuss here - and I am not sure if I can provide the answer to all of them without knowing the details of the technical implementation of the tracking in the study and the raw data, but I will try to provide my best guesses.
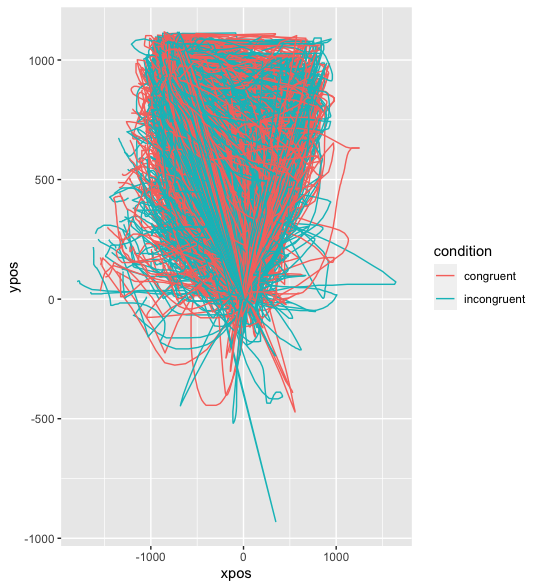
1) Upside down raw trajectories
In many mouse-tracking implementations it is the case that in the raw data the y coordinates increase as the cursors moves downwards, so I think that this is fine. To flip the trajectories you could use mt_remap_symmetric if (and only if) the coordinate system is centered. If this is not the case, which I guess is true here, you could flip them manually, e.g., with the following command:
mt_data$trajectories[,,"ypos"] <- (-mt_data$trajectories[,,"ypos"])2) Implausible coordinates
I can only speculate about this question as it depends on the implementation of your study. How did you control the size of the window of your study if it was browser based? That is, is it really not possible for the user to increase the size of the browser window? And did you check what happens with the recorded positions if the cursor leaves the browser window?
3) Error and warning messages
I assume that the raw data are provided in long format and imported using mt_import_long. If so: are there any error messages already displayed at the step of the data import?
Best,
Pascal
Regarding the third point: I just saw your post in the other thread here: http://forum.cogsci.nl/index.php?p=/discussion/5744/mt-aggregate
Given that there are warnings already at data import, I think that these are the cause of some of the errors and warnings happening later. We can discuss in the other thread how to fix the data import.
Hi P_Barr,
we are also working on importing the output of Mathur & Reichling into Mousetrap but we are having some problems with it (e.g. reordering the stimuli without the key). Could you tell us how you modified the code?
Thanks in advance!