Bayesian ANCOVA
Hello team,
I am trying to write up the results for a Bayesian ANCOVA. As there are no specific instructions on conducting/reporting this in the manual, I am facing some issues in ensuring if whatever I am writing makes sense.
Here is my output:
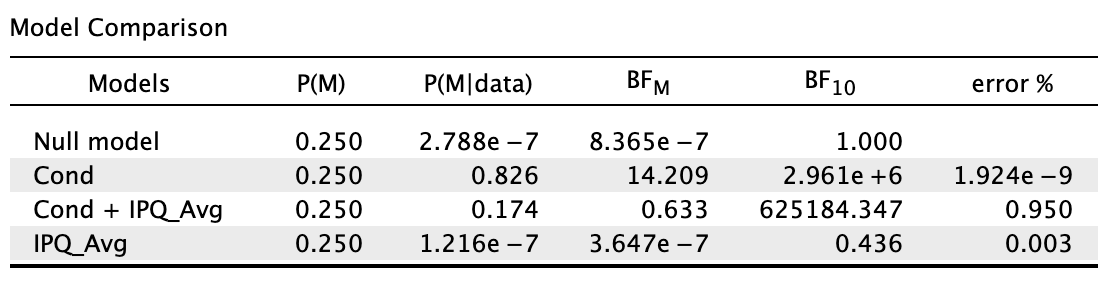
In short, cond = my IV and IPQ_Avg = my covariate.
Am I right to say this: A Bayesian ANCOVA was conducted, where a model containing test condition and sense of presence was compared against the null model containing only sense of presence. The default JASP priors (r scale prior width = 0.5 for fixed effects, r scale prior width = 0.354 for covariates) were used. Given the transitive relationship between Bayes factor, the model with test condition and sense of presence (BF10 = 625,184.35) was compared to the sense of presence model (BF10 = 0.44) by division (625,184.35/0.44 ≈ 1,420,873). Hence, after explaining for the error variance attributable to sense of presence, there was still extreme evidence for the effects of test condition on object location memory despite it being weaker."
My primary concern with this write-up was if my interpretation was correct, by comparing 1,420,873 (after controlling for presence) to 2,961,000 (without controlling for presence) and showing that there is still extreme evidence for the effects of test condition. Am I doing the transitivity thing correctly?
Thanks a ton.


Comments
The easiest way to check whether you did this correctly is to add "sense of presence" to the null model (under the tab "Model"). You might also want to tick "compare to best model" and "BF01" to the entries in the BF column tell you how much more likely the data are under the best model than any of the others.
Cheers,
E.J.
Hi E.J.,
Here's my output:
Am I right to say that the cond model is the best model and adding presence into the model weakens it? However, my concern here is whether I should be comparing cond + presence model with presence model (as per what I wrote in the original post above) or cond model with cond + presence model in controlling presence as a covariate.
Please do advise, thank you so much E.J.
Yes.
Second, "my concern here is whether I should be comparing cond + presence model with presence model (as per what I wrote in the original post above) or cond model with cond + presence model in controlling presence as a covariate."
If you want to assess whether "presence" adds anything over and above the cond model ("should the covariate be added?"), you compare cond to cond+presence. If you want to see whether there is evidence for cond while controlling for the covariate, you compare presence to cond+presence.
Cheers,
E.J.
Hi E.J.
I am new to Bayesian analysis. I am wondering what is the actual effect of adding a covariate to the null model (under the tab "Model") as you suggested in this thread. is it the same as treading them as a nuisance regressor?
I am analyzing cortical thickness differences between two groups (patients vs control) and I want to treat age, head motion during imaging, and MRI ghosting as nuisance regressors. Should I add these covariates to the null model for a Bayesian ANCOVA?
Best,
Luis
Yes. But I do think it is important to check that these nuisance factors do not differ between the groups. Suppose that cortical thickness is lower for older people, and patients are older than the controls. Instead of removing noise, you may then be removing signal. Not sure whether there exists a good solution in such cases (because there is an inherent confound) but it is good to be aware.
Cheers,
E.J.
Thank you E.J.
My cohorts are age- and sex-matched. With similar measurements for efc (MRI ghosting artifact). Here are the Bayesian T-Test for age and efc:
and the chi-squared test for sex:
Here is the output of the Bayesian ANCOVA for the thickness of the left paracentral gyrus (normalized to the thickness of an area not expected to change as a result of the condition, as suggested by the freesurfer team) when adding age and efc to the null model.
Is it accurate to describe these results as follows?
We performed a Bayesian analysis of covariance on the thickness of the left paracentral gyrus (normalized to the thickness of the left lateral precentral gyrus), including spinal cord injury diagnosis (yes, no) as a fixed factor, with efc (an empirical measurement of MRI ghosting artifacts) and age as nuisance factors.
An alternative model containing a spinal cord diagnosis as a predictor of cortical thickness was compared against a null model. All models contained efc and age as nuisance factors. The default JASP priors were used with both models set to be equally likely (P(M) = 0.5). After observing data, only the alternative model had its model odds increased (BFm = 12.858).
These results strongly suggest that the observed data is 12.8 times more likely under the alternative model. On average the grey matter of the area somatotopically representing the right leg and foot (left paracentral gyrus) of participants living with a spinal cord injury is 91.5% as thick as the area of their brains representing the face (left lateral paracentral gyrus) with a 95% credible interval of 88.88% to 94.16%. On the other hand, the left paracentral gyrus of participants without a spinal cord injury is on average 96.28% (CI = 94.63 - 97.91) as thick as their left lateral precentral gyrus.
I would greatly appreciate any input you may have.
Best,
Luis
Hi Luis,
This sounds reasonable. I would replace "These results strongly suggest that the observed data is 12.8 times more likely under the alternative model." to "These results show that the observed data is 12.8 times more likely under the alternative model."
Cheers,
E.J.
Thank you again for taking the time to answer.
I have one more follow up question. I have 2 other cortical regions I am analyzing. After running the ANCOVAS, should I correct for multiple comparison (3 regions of interest * 2 hemispheres)? if so, is there a method in JASP to do so?
Best,
Luis
Dear Luis,
The correction goes by means of the prior model probability. See for instance this summary:
Cheers,
E.J.
E.J.
Thank you very much for referring me to that paper. It was very informative.
I am a bit confused about the prior model probability I should use for my comparisons when trying to implement the null control (either Jeffreys or Westfall). According to the manuscript you shared with me, for this type of corrections with 4 groups the priors calculation would look like so (page 9):
In my case I have 6 means for the thickness of 6 different regions of interest that i am comparing across 2 groups using 6 ANCOVAS (with age and head movement as nuisance). I do not think I should use m= 6 (ROIs) since that would be k = 15 and I am not doing 15 comparisons since my ROIs means are not being compared to one another. I also do not think I should use m = 2 (groups) since that is the same as not doing any corrections (k = 1).
Can I set k = 6 (since I am doing 6 distinct comparisons across 2 groups) and use a Westfall prior of 0.707 to account for dependency?
Thank you again for all the help and guidance you have provided thus far.
--Luis
Makes sense to me!
Cheers,
E.J.
Great!
Thank you.
Hi EJ.
Reading through some other posts, I think I confused myself about correcting for multiple comparisons. I have a few questions pending:
You'd have to add that prior model probability yourself. We have it implemented under our ANOVA posthoc tests, but that is a specific scenario that does not apply exactly here.
Once you have the prior model probability p, you transform that to a prior odds p/(1-p) and multiply this by the Bayes factor to get your posterior odds.
So yes, you need to do the update manually.
E.J.
Hello,
thanks for discussing the topic on multiple comparisons;
Could you break the correction again down for me step by step? I have, as Luis, 6 hypotheses that I am testing in 6 different bayes ANCOVAS. How can I correct for multiple comparisons?
thanks! Birgit
I'd go over https://psyarxiv.com/s56mk
Ultimately this is about the prior plausibility of the hypotheses.
E.J.