Why does Partial least squares SEM perform slow?
Hi,
Thank you for providing PLS-SEM function in JASP. I really like it. But I have confronted a situation that really bother me a lot. When I finished setup all the parameters (see the attached) and start analysis, the speed of analysis is really slow. I can't figure out what is the problem. Any idea? My model specification is as follows and the data is also attached:
# measurement model
UB =~ UB1 + UB2 + UB3 + UB4
VB =~ VB1 + VB2 + VB3 + VB4
PR =~ PR1 + PR2 + PR3 + PR4 + PR5 + PR6 + PR7 + PR8
FR =~ FR1 + FR2 + FR3 + FR4
TB =~ TB1 + TB2 + TB3 +TB4
IB =~ IB1 + IB2 + IB3 + IB4
PS =~ PS1 + PS2 + PS3
BI =~ BI1 + BI2 + BI3
# regressions
BI ~ UB+ VB + PR + FR + TB + IB + PS


Comments
Hello,
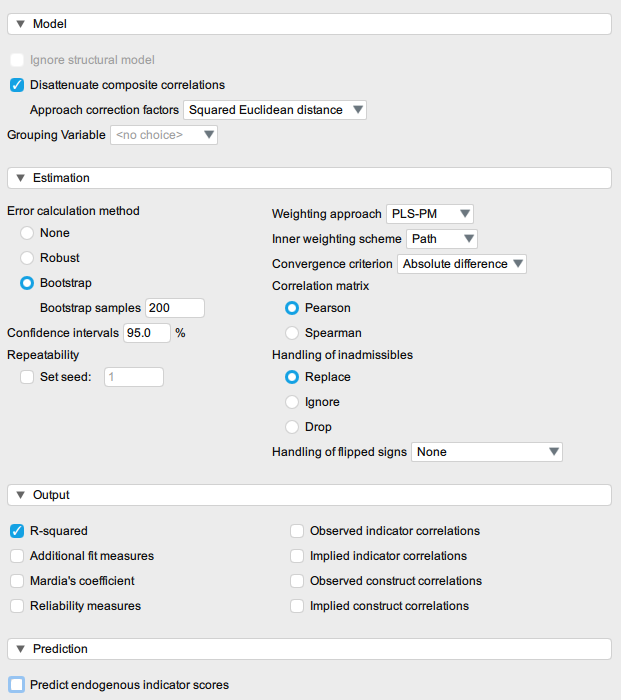
I can think of two reasons your analysis is slow: (1) You chose the bootstrap option in the estimation options, that means the analysis runs on 200 data sets, given that the number of samples is specified as 200 in your screenshot; changing the option to "none" will speed up the analysis, but it also means you won't be provided with confidence intervals. (2) Your data in JASP is scaled as "ordinal" which also slows down the analysis. If you wish to treat it as quasi-continuous you can change the scale of the data (https://jasp-stats.org/getting-started/, "Changing Variable Types").
Best,
Julius
Dear Julius,
Thank you for you quick answers. I have changed the scale of the data into scale type, but the slow situation doesn't improve. Regarding bootstrapping, I used to use R packages such as plspm and cSEM to perform PLS-SEM with the same dataset. With the same 200 or even 500 bootstrapping, it is quite obvious that plspm and cSEM can finish the analysis more quickly than JASP's PLS-SEM. I enjoy using JASP, so I really want to figure out what's the problem. Thank you very much for the answers.
Best wishes,
K.M.
Hmm, I agree that JASP will likely be slower than cSEM in R given that there is a bit of overhead with any analysis. I wonder if you can quantify the difference? Or might you be able to share the dataset so I can look into it?
Best,
Julius
Dear Julius,
I test the dataset with cSEM with 500 bootstrapping, it can finish in less than 11 secs.
R codes:
--------------------------------------------------------------------------------------
library(cSEM)
# initialization
#------------------------------------------------------------------------------
rm(list = ls())
encoding = "UTF-8" # To avoid warning message
# read data
file = "C:\\temp\\Data for PLS-SEM.csv"
D2P <- read.csv(file)
#-------------------------------------------------------------------------------
# setup model structure
#-------------------------------------------------------------------------------
# setup pls model
pls.model <- "
# Structural model
BI ~ UB + VB + PR + FR + TB + IB + PS
# Reflective measurement model
BI =~ BI1 + BI2 + BI3
UB =~ UB1 + UB2 + UB3 + UB4
VB =~ VB1 + VB2 + VB3 + VB4
PR =~ PR1 + PR2 + PR3 + PR4 + PR5 + PR6 + PR7 + PR8
FR =~ FR1 + FR2 + FR3 + FR4
TB =~ TB1 + TB2 + TB3 + TB4
IB =~ IB1 + IB2 + IB3 + IB4
PS =~ PS1 + PS2 + PS3
"
start.time <- Sys.time()
pls.result <- csem(.data = D2P,
.model = pls.model,
.approach_paths = 'OLS',
.disattenuate = FALSE,
.approach_weights = "PLS-PM",
.PLS_approach_cf = "dist_squared_euclid",
.PLS_weight_scheme_inner = 'path',
.handle_inadmissibles = "replace",
.resample_method = "bootstrap",
.R = 500,
.seed = 123,
.iter_max = 100)
end.time <- Sys.time()
# May conflict with dplyr
(summary_pls_result <- cSEM::summarize(pls.result))
(time.taken <- end.time - start.time)
---------------------------------------------------------------------------------------
But the JASP's PLS-SEM with 200 bootstrapping starting at 21:00, you can see the progress bar at 21:07. There seems a long way to go.
21:00
21:07
Please see the attached for the data file.
Thanks for you help.
Best wishes,
K.M.
you are correct. It is quite slow in JASP. I will look into it. May I have your permission to transform this into a GitHub issue?
Dear Julius,
You have my full permission to transform this into a GitHub issue. Thank you very much for you help.
Best wishes,
K.M.
Ah, so that was longer detective work than I anticipated, given that the issue is kind of obvious :) But in short, the issue comes from setting "disattenuate = TRUE". In your R code you have disattenuate = FALSE, which makes the code run smooth because the resampling somehow does not produce inadmissible results. In JASP you have the checkbox for disattenuation checked, meaning it is set to TRUE, and then cSEM produces inadmissible results, and a lot, and having that handling set to "replace" means it is resampled A LOT. I dont really know how the disattenuation actually affects this, but it does. However, since the same happens in R with cSEM this is not a bug.
Best,
Julius
And given that there was this bug https://github.com/jasp-stats/jasp-issues/issues/2260 you might not reproduce this as the checkbox for disattenuation has no effect in the current JASP version; it is always set to TRUE.
Dear Julius,
Thank you for solving my question.
K.M.