Quasi randomization of trials method
Hi there,
I'm currently exploring Open Sesame and OSWeb as a way of running experiments online, trying to work out whether it can handle the specific requirements my experiments typically have. One of these requirements is the generation of list of trials following sometimes demanding quasi randomization conditions (e.g., making sure that not more than two trials of such or such condition are presented consecutively, that such or such stimulus is not repeated on consecutive trials etc.). I use such quasi randomization to create a unique trials list order for each participant. So far I've done my experiments using E-Prime, using EBasic to write code, or when the quasi randomization is so complex that the program can get stuck without a solution, I've written an application capable of batching stimuli files that fulfill all the conditions, and then have the task import these, commit all information to variable arrays and then populate the trials list at the onset of the experiment.
I'm new to OpenSesame and OSWeb, and so far I haven't been able to work out how to achieve a similar result. I thought of the following options and have doubts or questions related to each:
(1) Quasi randomization through code:
I understand that Python code could take care of randomization, but this would not be suitable to run the experiment online as OSWeb does not integrate python code objects, so I guess that rules out Python code. As for Javascript code, I understand that, within Open Sesame, it is restricted to defining and reading from variables. Is that correct? (if it is, then I guess it rules a Javascript solution too). Or can OSWeb handle javascript containing for, do/while loops and some functions such as Math.floor, Math.random, and array manipulations? Is there a method to achieve my aim through coding that would work in OSWeb?
(2) Reading from text file and populating a loop item table
Is there a way to read the content of a text file (e.g., csv), to commit it to memory arrays and then populate a loop item table with that information? For example, reading stimulus and correct response information for 20 trials from a text file, and then populate a 10 trials loop item table with information for trials 1 to 10 on the first execution of the loop, and with information from trials 11 to 20 on the second execution of the loop?
(3) Importing trials information from text file into a loop item dynamically
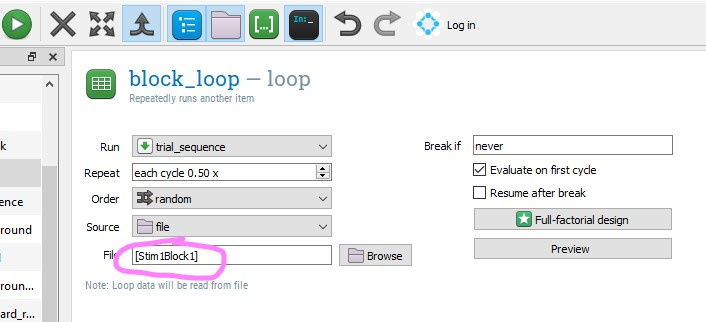
I've seen that the trials list / loop item can be populated from a file instead of a table. Can the path and file name be specified dynamically? For example, setting up variables such as StimFileBlock1 = "stim" + subect_nr + "B1.csv"; StimFileBlock2 = "stim" + subect_nr + "B2.csv"; etc. And then dynamically using these variables to specify the file to import in the loop item...
Sorry for the rather long message... I hope what I wrote is complete enough to explain the different options I've considered. Any help would be much appreciated on these or indeed or any alternative solution.
Thanks in advance for any advice!
Best,
Fabrice.



Comments
Hi @Fab ,
There are indeed several options to apply pseudorandomization, and you already figured out most of them:
Firstly, some basic constraints (minimum distance and maximum number of repetitions) can be applied directly in the
loopitem itself:Secondly, for more complex constraints, you could use some Python script in
inline_scriptitems, for example the package pseudorandom:And for more info on Python inline scripting:
Finally, you could indeed prepare the trial lists in advance and read them in with a
loopitem. Yes, you can make the name of the file variable (for example depending on the participant number) by using thesquare-bracket syntaxlike so:(Note that 'subject_nr' is a variable that is automatically created by OpenSesame, so it always exists.)
For more information on the square-bracket syntax, see here:
Hope this helps!
Cheers,
Lotje
Did you like my answer? Feel free to

Hi Lotje,
Many thanks for these pointers!
Solutions 1 and 3 should solve my problem (I don't think Solution 2 would as, if I understand correctly, python inline_script is not supported by OSWeb and so it would not be adequate for running the experiment online. Is that correct?).
I've just tried solution 1 with a simple condition and it works really well. My concern is what might happen for complex conditions where the program might hang and crash. Solution 3 would certainly be ideal in that case, though to run the experiment through OSWeb, using the subject ID number to select a text file with the trials list wouldn't be an option since the task is running on the client machine and so it can't know how many participants have already taken part to use an incremental subject number procedure. I guess that in that case, I could just create a large set of text files (say 500) and select a value at random for each subject using an inline_javascript object (vars.fileid = Math.floor((Math.random() * 500) + 1).
Many thanks!
Fabrice.
I think what @lvanderlinden was trying to say is that you can also use java inline scripting to achieve the same thing ;)
You can use while and for loops and math functions in the java codeblock, no problem. It might not work sometimes within OS, but work fine when you run it in the browser (so that should be your test). When you code the pseudorandom variables, make sure to save them to a vars.[variable_name] variable (the vars part ensures that you can access this variable later on, as opposed to just using var).
Good luck!
Hi @JasperdeWaard,
Thanks! I'll read up on Javascript (never used it until now). I think that in the short term I'll prepare csv files with the information for the loops and select a few at random to upload them for my different blocks.
I'm trying to do this dynamically, using javascript to pick random numbers, which I'm then saving in an array (fileid[]). Later on, I'm using a loop variable (nb) to retrieve the content of fileid[nb], which I save into a stimfile variable, which I use to specify which csv file my tets_trials loop should import. It's working when I execute it in an Open Sesame window, but it fails if I try to run it in my browser. I think it is because it's failing to handle the variables as in Open Sesame and failing to load up a csv file in the test_trials loop, it reverts to the table option, which is empty. I've been going in circle for hours now. I'm making very little use of Javascript, so I was hoping it would run well in a browser. There must be something I'm missing...
I'm attaching some screen capture in case you or anybody reading this is inspired and can help me out.
Best,
Fabrice.
Hi @lvanderlinden and @JasperdeWaard,
Following my message above, I worked out that the problem is not to do with the variables or bits of javascript I'm using. Instead, the problem is that for some reason, the program ignores the source of my loop. While I set it to use a file, it ignores it and runs it as a table. However, it only does that when executed in a browser. If I execute the program within a window, it runs just fine.
So I created a new task, the most basic possible, just to test the file source in OSWeb. No javascript, no variables, just setting up the source as List1.csv. Runs fine in Open Sesame but fails in the browser.
Could it be that the file source for loops is not supported in browser or through OSWeb? Or have I missed something?
Thanks in advance for any help!
Fabrice.
Hi @Fab ,
My apologies, as you noticed, in my first answer I didn't quite get that you were asking specifically for pseudorandomization online. Unfortunately, no, reading files is indeed not supported (yet) in OSWeb...
I'll give your issue a thought...
And thanks for being so active on the forum! :)
Cheers,
Lotje
Did you like my answer? Feel free to

Hi @lvanderlinden,
Thanks for getting back to me! That compatibility issue had escaped my attention. Would have saved you time if I had paid more attention to that web page! Unlucky for me that file sources are not supported when running the task in a browser. That'd be a great feature to add to OSWeb in the future.
For the experiment I'm planning to run, that's a critical limitation. To guarantee some variety in the trials lists across subjects, I currently randomly pick a trials list from a collection of 200 csv files. The ordering of the trials is quite complex and I think it'd take me a long time to learn enough javascript to program the pseudorandomization within the task. Might give it a go eventually (learning some Javascript is on my bucket list), but in the mean time I think I'll look into alternative programs (started looking into JsPsych and PsychoPy) even if I think that Open Sesame is really great (especially in combination with the Mindprobe initiative)!
Thanks again for your help! :-)
Cheers,
Fabrice.
@lvanderlinden,
PS: Hi again, Lotje... Just read your posts on https://forum.cogsci.nl/discussion/6978/randomly-select-some-trials-from-two-distinct-groups (great stuff!) and it got me thinking... I saw that you use javascript to define arrays (in the prepare phase of the experiment), and that some code can then be used to pick one at random and populate dynamically the loop before it runs. I suppose I could adapt my stimulus lists generator (some application I wrote in Visual Basic) to output them in a javascript format I'd then just have to paste in an inline javascript object in Open Sesame. But I wonder whether it would be reasonable in my case, for I'm talking about say 200 lists x 1216 trials x 3 columns. I'm a little worried it might take up too much memory and make the task sluggish. What do you think?
Hi Fab,
I hope you don't mind if chime in here. 200x1216x3 sounds like a lot. I don't think it necessarily would cause an issue with memory, especially because you don't need to carry the full thing all the way through the experiment. So after you created the complete list, you can select the one that you need for your participant, and deleting the rest, then you would end up with only 1216x3, which is definitely no problem.
I guess you should just try and see whether it works?
Alternatively, you don't need to always create 200 lists. Perhaps, you can split the same experiment in 4-8 separate experiments, that are identical except that a different pool of 25-50 x1216 x 3 lists will be created. That could also reduce the memory load.
I had another thought on how to bypass the restriction of loading a file to Osweb. Perhaps you could upload the file as some sort of website. If the file content is text, OSweb might be able to read out the website and use the text information in the experiment. No clue, whether this makes sense, as I don't know enough about OSweb and javascript. @sebastiaan, @lvanderlinden would that be possible?
Eduard
Hi @eduard ,
Thanks for your input! Hadn't thought of deleting the arrays I don't need. Great idea! I think that then the memory demand should be quite small. After all, I'd just have one 1216 x 3 array containing text elements.
The idea of splitting the experiment in several experiments is good too. I could have my online subjects landing on some page that randomly redirects them to one of the versions of the experiment. But I think the first solution is best (I always try to keep everything in one task to avoid errors if some bits have to be updated).
The third solution sounds interesting too! If there was some way of reading the content of some external file into an array, I could select a file among 200 at random, and then only read that one. Solution 1 is probably the simplest for me at this stage, but I'll look into it just for the fun of it! (I had never touched Javascript until 5 days ago and my brain is near meltdown 😉).
Thanks a lot for the suggestions!
Cheers,
Fabrice.
@eduard @Fab
I had another thought on how to bypass the restriction of loading a file to Osweb. Perhaps you could upload the file as some sort of website. If the file content is text, OSweb might be able to read out the website and use the text information in the experiment. No clue, whether this makes sense, as I don't know enough about OSweb and javascript.
In principle, JavaScript can read text files from a URL (with
fetch()). However, this is usually forbidden by the so-called same-origin policy, which holds that you cannot read from other domains.So I think that loading all the data into one gigantic
inline_javascriptis indeed the easiest solution. Another solution would be to use JATOS's batch-session-data functionality to keep the different lists on the server, and request only the one that you want to use. That's much more complicated though, and I would first try the easy route!— Sebastiaan
Check out SigmundAI.eu for our OpenSesame AI assistant!
Hi @sebastiaan, (cc @eduard & @lvanderlinden )
Thanks for your thoughts on this! I did try to play around with the fetch function and looked up all kinds of functions but came up blank. Found some funky and straightforward Javascript code but to allow a client user to upload a csv file using the HTML input method, but never worked out how to replace that input by a path on the server.
Anyway, long story short, I'm pushing ahead with the long inline_javascript. I adapted my stimuli list generator in Visual Basic to output a single text file with all lists written in a javascript format that I then just have to paste in OS. I followed Eduard's suggestion to empty the arrays not used to free memory. I'm still working on it but when I've got it working, I thought I could annotate it and post it on the forum with some description in case it can be useful to other users.
Thanks again for your input!
Cheers,
Fabrice.
Sounds like a plan 👍️
Hi there,
I've just posted the method I've been working on in a new discussion thread: https://forum.cogsci.nl/discussion/7081/a-method-to-randomly-select-a-stimulus-list-for-online-experiments-javascript-jatos-compatible#latest
Cheers,
Fabrice.